
We use the symbiosis between the benthic Hawaiian bobtail squid, Euprymna scolopes, and the bioluminescent bacterium, Vibrio fischeri, as a model to study the effects of beneficial bacteria on animal host tissues. Our immediate goals are centered on understanding the molecular mechanisms by which specificity is ensured during initiation and maintenance of the symbiosis and defining interactions between components of the host’s innate immune system and symbiotic and non-symbiotic bacteria. Understanding how the innate immune system interacts with beneficial bacteria is an emerging area of interest in a number of fields of biology. Because invertebrates lack an adaptive immune system (meaning they have no antibody response), it is unclear how the innate immune system of this group of animals mediates highly specific recognition responses to microorganisms. The squid-vibrio association offers an experimentally tractable model to explore this question and our studies are setting the groundwork for understanding interactions between the symbiont and the innate immune system of the host.

We previously showed that host hemocytes can differentiate between symbiotic and non-symbiotic bacteria and that colonization state alters hemocyte binding of V. fischeri specifically (Nyholm et al., 2009). To understand the molecular basis of the host’s hemocyte response, we recently characterized the transcriptome and proteome of circulating blood cells (Collins et al., 2012) and showed that colonization of the adult light organ can influence the hemocyte proteome (Schleicher et al., 2014). We have described expression of a number of innate immunity genes and proteins including a novel peptidoglycan recognition protein, EsPGRP5. PGRPs are pattern recognition receptors (PRRs) often involved with the recognition of microbe-associated molecular patterns (MAMPs). We are currently characterizing the role of this and other PRRs in the squid-vibrio symbiosis. We are also using quantitative transcriptomic and proteomic techniques to characterize hemocytes from different stages of colonization and in response to challenge from symbiotic and non-symbiotic bacteria.
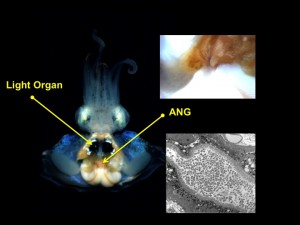
We are also developing the Hawaiian bobtail squid as a model host for the investigation of a second bacterial symbiosis found only in female squid. The accessory nidamental gland (ANG) is a reproductive organ that contains a bacterial consortium hypothesized to be involved with protection of the developing embryos in externally laid eggs from either fouling and/or predation. We have used a number of molecular techniques to characterize the members of the bacterial community and show that different bacterial groups partition to different portions of the ANG. We further demonstrated that bacteria from the ANG are deposited directly into the egg cases (Collins et al., 2012; Appl. Environ. Microbiol). We have a number of the bacterial members in culture and are currently sequencing the genomes of these. We are also characterizing bacterial gene expression in the eggs and ANG, experimentally altering the microbial community of the eggs with antibiotics to further understand the function of this association, and characterizing the development of the ANG and its bacterial consortium. We are currently collaborating with the Balunas Lab in the department of Pharmaceutical Sciences at UCONN to understand the function of these bacteria in the symbiosis.
 Hydrothermal Vent Symbioses
Hydrothermal Vent Symbioses
Our laboratory also investigates the molecular interactions between hydrothermal vent tubeworm hosts and their chemoautotrophic bacterial symbionts. Some of the highest primary productivity rates to date have been measured in these associations, yet the molecular mechanisms regulating host cell/symbiont interactions and metabolite function are almost entirely unknown. We are currently investigating issues of host/symbiont gene expression in collaboration with Dr. Peter Girguis of Harvard University. We have compared gene expression between the organ that house the symbiotic bacteria (trophosome) and the organ that interfaces with the environment (plume) to better understand host genes that may help regulate the symbiosis (Nyholm et al., 2012, PLoS ONE). We analyzed expression of a number of innate immunity genes under in situ conditions and found significantly higher expression of host pattern recognition receptors that potentially recognize microbe-associated molecular patterns (MAMPs). Our work supports the hypothesis that a molecular “dialogue” between the partners, including components of the host’s innate immune system, leads to a well-regulated association.